Single Cell Scheduler
Jump to navigation
Jump to search
Contents
Contact Information[edit]
- All inquiries about 10x Single Cell should be directed to ngsequencing@illinois.edu.
Important Information[edit]
- Timing is critical. Please contact the DNA Services lab, Chris Wright and Alvaro Hernandez at ngsequencing@illinois.edu to schedule your single cell experiment BEFORE you begin growing cells to ensure availability for processing.
- In order to set up and execute your test counts and/or single cell experiment properly, we need this form(10x_Single_Cell_Form), filled out completely. Email form to us at least 24-hours before your 10x experiment date.
- Starting cell concentration should be at least 4,000 cells/ul (4M cells/ml) or more. We request 1M cells total, transported to us in a 1.5ml centrifuge tube. There are modified protocols for limited cells, please ask if this is your case.
- Elimination of dead cells is critical, viability should be >70%. Your starting cell suspension will be counted upon arrival with live/dead measure included. If viability is below 70%, you may wish to repeat cell collection as background RNA levels will be high.
If cells cannot be obtained with >70% viability, 10x recommends the Miltenyi Biotec Dead Cell Removal kit with MS columns (catalog # 130-090-101 / 130-042-201). The magnet and stand for these columns can be checked out from the Flow Cytometry facility: https://biotech.illinois.edu/flowcytometry - Prior to library setup, you will need to tell us below how many cells should be targeted for each library, and how many different samples you have. A library can be made from 500-10,000 cells, depending upon your project needs. Doublets increase linearly with total cells, from ~0.6% at 1000 cells, to ~6% at 10,000 cells.
**Recommended target per sample is 3,000 cells or more. While libraries can be made to target 500-10,000 cells each, libraries capturing < 3,000 cells have much greater variability in actual cells captured (+/- 50% or more) and lower reproducibility. Variability of actual capture at or above 3,000 cells should be ~30% or less (ie: 2,700-3,300 for a 3,000-cell target). While all protocols for library preparation will be followed, due to differences in sample types submitted for 10x single cell, no guarantee is given that your cell type will behave according to typical performance. - For V3 single cell 3’ kits, 10x recommends at least 50k sequencing reads per targeted cell. At the 50k target, you can sequence 2 libraries of ~4000 cells in 1 lane of the NovaSeq SP (~400M single reads/lane). Once your need reaches 3 lanes of SP, cost is similar to 1 lane of NovaSeq S4 2x150nt and and S4 lane is recommended.
- Costs and sequencing options:
- Single library: $2,130; 2-7 libraries at the same time: $1,930 each; and 8+ are $1,810 each.
- Per lane NovaSeq SP 2x150nt: $2,720. ~400M 10X RNA reads per lane
- Per lane NovaSeq S1 2x100nt: $4,890. ~800M 10X RNA reads per lane
- Per lane NovaSeq S1 2x150nt: $5,270. ~800M 10X RNA reads per lane
- Per lane NovaSeq S4 2x150nt: $8,990. ~2.5M 10X RNA reads per lane
Using Online Scheduler to Schedule Meetings, Test Counts, and Experiments[edit]
10x Single Cell Scheduler - https://www-app.igb.illinois.edu/singlecell/
Gaining Access to Online Scheduler[edit]
Email this form(10x_Single_Cell_Scheduler_Access_Form) to ngsequencing@illinois.edu. You will receive an email when you have access to login to the scheduler. This scheduler is only used to schedule BL1 test counts. Anything else must be scheduled by contacting ngsequencing@illinois.edu.
Scheduling an Initial Meeting[edit]
- To schedule an inital meeting, please contact ngsequencing@illinois.edu
Scheduling Test Counts[edit]
- There is no charge to run test counts.
- Any new cell type should have at least one successful test count prior to the full experiment.
- Plan your test counts! Test count and full experiments must be scheduled 1 week in advance. The calendar will not allow you to schedule within 7 days of present time.
- Make sure you schedule the appropriate amount of time for the number of samples you have.
- 1-2 samples - 1 hour
- 3-4 samples - 1 hour and 30 minutes
- 4+ samples - 2 hours
- Test counts for BL1 are performed in ERML 221.
- If your samples are BL2, you MUST contact ngsequencing@illinois.edu to schedule test counts!!
- Login to the scheduler using your netID and AD password.
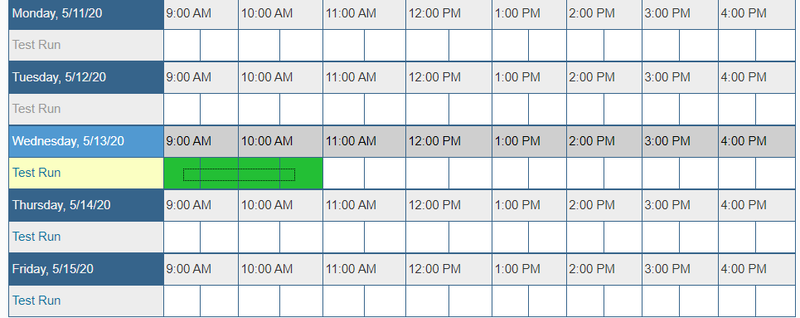
- Next to "Test Run", click on the time you would like to schedule a meeting.
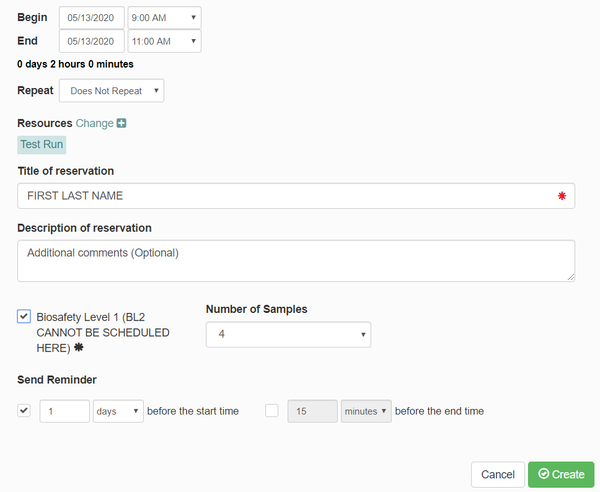
- A box will pop up to make your reservation. Enter the following information:
- Verify the date and time of your meeting
- Under "Title of reservation", enter your first and last name.
- Under "Description of reservation", enter any additional comments you believe are needed.
- Under "Number of Samples", enter the number of samples you will be using (you can have a maximum of 16 for one experiment).
- Click "Create"
- You will receive confirmation of your reservation and should be able to see it on the calendar now.

Scheduling the Experiment[edit]
- Contact ngsequencing@illinois.edu to schedule your experiment.