Difference between revisions of "Biocluster"
(→On Mac OS X) |
(→How To Submit A Cluster Job) |
||
| (25 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__TOC__ | __TOC__ | ||
| + | ==Quick Links== | ||
| + | * Main Site - [http://biocluster.igb.illinois.edu http://biocluster.igb.illinois.edu] | ||
| + | * Request Account - [http://www.igb.illinois.edu/content/biocluster-account-form http://www.igb.illinois.edu/content/biocluster-account-form] | ||
| + | * Cluster Accounting - [https://bioapps3.igb.illinois.edu/accounting/ https://bioapps3.igb.illinois.edu/accounting/] | ||
| + | * Cluster Monitoring - [https://bioapps3.igb.illinois.edu//ganglia/ https://bioapps3.igb.illinois.edu/ganglia/] | ||
| + | * SLURM Script Generator - [http://www-app.igb.illinois.edu/tools/slurm/ http://www-app.igb.illinois.edu/tools/slurm/] | ||
| + | * Biocluster Applications - [https://help.igb.illinois.edu/Biocluster_Applications https://help.igb.illinois.edu/Biocluster_Applications] | ||
| + | * Biocluster Introduction Presentation - [https://help.igb.illinois.edu/images/d/df/Intro_to_Biocluster_Spring_2022.pptx Intro_to_Biocluster_Spring_2022.pptx] | ||
| + | ==Description== | ||
| + | Biocluster is the High Performance Computing (HPC) resource for the Carl R Woese Institute for Genomic Biology (IGB) at the University of Illinois at Urbana-Champaign (UIUC). Containing 2824 cores and over 27.7 TB of RAM, Biocluster has a mix of various RAM and CPU configurations on nodes to best serve the various computation needs of the IGB and the Bioinformatics community at UIUC. For storage, Biocluster has 1.3 Petabytes of storage on its GPFS filesystem for reliable high speed data transfers within the cluster. Networking in Biocluster is either 1, 10 or 40 Gigibit ethernet depending on the class of node and its data transfer needs. | ||
| − | + | * The biocluster is not an authorized location to store '''HIPAA''' data. | |
| + | *'''If you need to update the CFOP associated with your account, please send an email with the new CFOP to help@igb.illinois.edu.''' | ||
| − | + | ==Cluster Specifications== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | {| border="1" class='wikitable' width="1200" cellspacing="1" cellpadding="0" align="center" |
| + | |- | ||
| + | !|Queue Name | ||
| + | !|Nodes | ||
| + | !|Cores (CPUs) per Node | ||
| + | !|Memory | ||
| + | !|Networking | ||
| + | !|Scratch Space /scatch | ||
| + | !|GPUs | ||
| + | |- | ||
| + | ||normal (default) | ||
| + | ||5 Supermicro SYS-2049U-TR4 | ||
| + | ||72 Intel Xeon Gold 6150 @2.7 GHz | ||
| + | ||1.2TB | ||
| + | ||40GB Ethernet | ||
| + | ||7TB SSD | ||
| + | || | ||
| + | |- | ||
| + | ||lowmem | ||
| + | ||8 Supermicro SYS-F618R2-RTN+ | ||
| + | ||12 Intel Xeon E5-2603 v4 @ 1.70Ghz | ||
| + | ||64GB RAM | ||
| + | ||10GB Ethernet | ||
| + | ||192GB | ||
| + | || | ||
| − | + | |- | |
| + | ||gpu | ||
| + | ||1 Supermicro | ||
| + | ||28 Intel Xeon E5-2680 @ 2.4Ghz | ||
| + | ||256GB | ||
| + | ||1GB Ethernet | ||
| + | ||1TB SSD | ||
| + | ||4 NVIDIA GeForce GTX 1080 Ti | ||
| + | |- | ||
| + | ||classroom | ||
| + | ||10 Dell Poweredge R620 | ||
| + | ||24 Intel Xeon E5-2697 v2 @ 2.7Ghz | ||
| + | ||384GB | ||
| + | ||10GB Ethernet | ||
| + | ||750GB | ||
| + | || | ||
| − | + | |} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== Storage == | == Storage == | ||
| − | |||
=== Information === | === Information === | ||
| − | The storage system is a GPFS filesystem with | + | * The storage system is a GPFS filesystem with 1.3 Petabytes of total disk space with 2 copies of the data. This data is '''NOT''' backed up. |
| + | * The data is spread across 8 GPFS storage nodes. | ||
| − | === Cost === | + | ===Cost=== |
| − | {| class= | + | On April 1, 2021, CNRG was informed by campus that we were required to start billing external users paying with a credit card an external rate. This rate was given to us by campus and is obtained by adding the 31.7% F&A rate and adding the standard 2.3% credit card fee. This external rate is only charged to users paying with a credit card. |
| + | {| border="1" class='wikitable' width="600" cellspacing="1" cellpadding="0" align="center" | ||
|- | |- | ||
| − | ! Cost (Per Terabyte Per Month) | + | !|Internal Cost (Per Terabyte Per Month) |
| + | !|External Cost (Per Terabyte Per Month) | ||
|- | |- | ||
| − | | $ | + | ||$8.75 |
| + | ||$11.73 | ||
|} | |} | ||
| − | == | + | === Calculate Usage (/home) === |
| + | * Each Month, you will receive a bill on your monthly usage. We take a snapspot of usage daily. Then we average out the 95 percentile of daily snapspots to get an average usage for the month. | ||
| + | * You can calculate your usage using the du command. An example is below. The result will be double what you are billed as their is 2 copies of the data. Make sure to divide by 2. | ||
| + | <pre>du -h /home/a-m/username</pre> | ||
| − | The | + | == Calculate Usage (/private_stores) == |
| + | * These are private data storage nodes. They do not get billed monthly. | ||
| + | * The filesystems are XFS shared over NFS. | ||
| + | * To calculate usage, use the du command | ||
| + | <pre>du -h /private_stores/shared/directory</pre> | ||
| − | + | ==Queue Costs== | |
| + | The cost for each job is dependent on which queue it is submitted to. Listed below are the different queues on the cluster with their cost. Although the service is billed by the second, the rates below are what it would cost per day to use a resource, so that it would be more easily understood. For standard computation, the CPU cost and the memory cost are compared and the highest is billed. For GPU bills the cost of the CPU or memory is added to that of the GPU. | ||
| − | {| class= | + | Usage is charge by the second. The costs listed below are what it would cost per day. The CPU cost and memory cost are compared and the largest is what is billed. |
| + | {| border="1" class='wikitable' width="1400" cellspacing="1" cellpadding="0" align="center" | ||
|- | |- | ||
| − | ! Queue Name | + | !|Queue Name |
| − | ! CPU Cost | + | !|CPU Cost |
| − | ! Memory Cost | + | !|External CPU Cost |
| + | !|Memory Cost | ||
| + | !|External Memory Cost | ||
| + | !|GPU Cost | ||
| + | !|External GPU Cost | ||
| + | |||
|- | |- | ||
| − | | default | + | ||normal (default) |
| − | | $1. | + | ||$1.19 |
| − | | $0. | + | ||$1.59 |
| + | ||$0.08 | ||
| + | ||$0.09 | ||
| + | ||NA | ||
| + | ||NA | ||
|- | |- | ||
| − | | | + | ||lowmem |
| − | | $ | + | ||$0.50 |
| − | | $0. | + | ||$0.67 |
| + | ||$0.10 | ||
| + | ||$0.13 | ||
| + | ||NA | ||
| + | ||NA | ||
| + | |||
|- | |- | ||
| − | | | + | ||GPU |
| − | | $ | + | ||$2.00 |
| − | | $ | + | ||$2.68 |
| − | + | ||$0.44 | |
| − | | | + | ||$0.59 |
| − | | $ | + | ||$2.00 |
| − | | $0. | + | ||$2.68 |
| − | |||
| − | | | ||
| − | | $ | ||
| − | | $ | ||
|} | |} | ||
| + | On April 1, 2021, CNRG was informed by campus that we were required to start billing external users paying with a credit card an external rate. This rate was given to us by campus and is obtained by adding the 31.7% F&A rate and adding the standard 2.3% credit card fee. This external rate is only charged to users paying with a credit card. | ||
| − | == | + | ==Gaining Access== |
| − | *Please fill out the form at [http://www.igb.illinois.edu/content/biocluster-account-form http://www.igb.illinois.edu/content/biocluster-account-form] to request access to the Biocluster. | + | * Please fill out the form at [http://www.igb.illinois.edu/content/biocluster-account-form http://www.igb.illinois.edu/content/biocluster-account-form] to request access to the Biocluster. |
| − | == Cluster Rules == | + | ==Cluster Rules== |
| − | *'''Running jobs on the head node are strictly prohibited.''' Running jobs on the head node could cause the entire cluster to crash and affect everyone's jobs on the cluster. Any program found to be running on the headnode will be stopped immediately and your account could be locked. You can start an interactive session to login to a node to manual run programs. | + | * '''Running jobs on the head node or login nodes are strictly prohibited.''' Running jobs on the head node could cause the entire cluster to crash and affect everyone's jobs on the cluster. Any program found to be running on the headnode will be stopped immediately and your account could be locked. You can start an interactive session to login to a node to manual run programs. |
| − | *'''Installing Software''' Please email help@igb.illinois.edu for any software requests. Compiled software will be installed in /home/apps. If its a standard RedHat package (rpm), it will be installed in their default locations on the nodes. | + | * '''Installing Software''' Please email help@igb.illinois.edu for any software requests. Compiled software will be installed in /home/apps. If its a standard RedHat package (rpm), it will be installed in their default locations on the nodes. |
| − | *'''Creating or Moving over Programs:''' Programs you create or move to the cluster should be first tested by you outside the cluster for stability. Once your program is stable, then it can be moved over to the cluster for use. Unstable programs that cause problems with the cluster can result in your account being locked. | + | * '''Creating or Moving over Programs:''' Programs you create or move to the cluster should be first tested by you outside the cluster for stability. Once your program is stable, then it can be moved over to the cluster for use. Unstable programs that cause problems with the cluster can result in your account being locked. Programs should only be added by CNRG personnel and not compiled in your home directory. |
| − | *'''Reserving Memory:''' | + | * '''Reserving Memory:''' SLURM allows the user to specify the amount of memory they want their program to use.. If your job tries to use more memory than you have reserved, the job will run out of memory and die. Make sure to specify the correct amount of memory. |
| − | *'''Reserving Nodes and Processors:''' For each job, you must reserve the correct number of nodes and processors. By default you are reserved 1 processor on 1 node. If you are running a multiple processor job or a MPI job you need to reserve the appropriate amount. If you do not reserve the correct amount, the cluster will | + | * '''Reserving Nodes and Processors:''' For each job, you must reserve the correct number of nodes and processors. By default you are reserved 1 processor on 1 node. If you are running a multiple processor job or a MPI job you need to reserve the appropriate amount. If you do not reserve the correct amount, the cluster will confine your job to that limit, increasing its runtime. |
| − | == How To Log Into The Cluster == | + | ==How To Log Into The Cluster== |
| − | *You will need to use an SSH client to connect. | + | * You will need to use an SSH client to connect. |
| + | *'''NOTICE''' The login hostname is '''biologin.igb.illinois.edu''' | ||
| − | === On Windows === | + | ===On Windows=== |
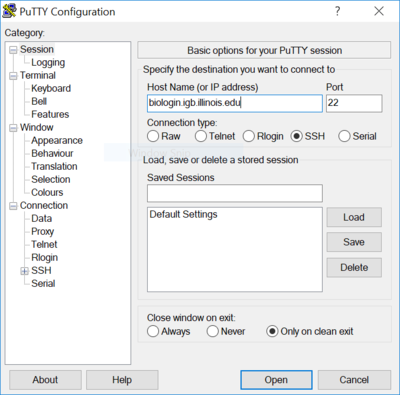
| − | *You can download Putty from [http://www.chiark.greenend.org.uk/~sgtatham/putty/download.html http://www.chiark.greenend.org.uk/~sgtatham/putty/download.html] | + | * You can download Putty from [http://www.chiark.greenend.org.uk/~sgtatham/putty/download.html http://www.chiark.greenend.org.uk/~sgtatham/putty/download.html] |
| − | *Install Putty and run it, in the Host Name input box enter '''biologin.igb.illinois.edu''' | + | * Install Putty and run it, in the Host Name input box enter '''biologin.igb.illinois.edu''' |
| − | |||
| − | + | [[File:PUTTYbiologin.PNG|400px]] | |
| − | + | * Hit Open and login using your IGB account credentials. | |
| − | + | ===On Mac OS X=== | |
| − | |||
| − | |||
| − | + | * Simply open the terminal under Go >> Utilities >> Terminal | |
| + | * Type in '''ssh username@biologin.igb.illinois.edu''' where username is your NetID. | ||
| + | * Hit the Enter key and type in your IGB password. | ||
| − | + | ==How To Submit A Cluster Job== | |
| − | |||
| − | |||
| − | |||
| − | === Create a Job Script === | + | * The cluster runs the '''SLURM ''' queuing and resource mangement program. |
| + | * All jobs are submitted to SLURM which distributes them automatically to the Nodes. | ||
| + | * You can find all of the parameters that SLURM uses at [https://slurm.schedmd.com/quickstart.html https://slurm.schedmd.com/quickstart.html] | ||
| + | * You can use our SLURM Generation Utility to help you learn to generate job scripts [http://www-app.igb.illinois.edu/tools/slurm/ http://www-app.igb.illinois.edu/tools/slurm/] | ||
| + | |||
| + | ===Create a Job Script=== | ||
| + | |||
| + | * You must first create a SLURM job script file in order to tell SLURM how and what to execute on the nodes. | ||
| + | * Type the following into a text editor and save the file '''test.sh''' | ||
| − | |||
| − | |||
<pre>#!/bin/bash | <pre>#!/bin/bash | ||
| − | # | + | #SBATCH -p normal |
| + | #SBATCH --mem=1g | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -n 1 | ||
sleep 20 | sleep 20 | ||
echo "Test Script" | echo "Test Script" | ||
</pre> | </pre> | ||
| − | + | * You just created a simple SLURM Job Script. | |
| − | + | * To submit the script to the cluster, you will use the sbatch command. | |
| − | *You just created a simple | + | |
| − | *To submit the script to the cluster, you will use the | + | <pre>sbatch test.sh</pre> |
| − | <pre> | + | * Line by line explanation |
| − | *Line by line explanation | + | ** '''#!/bin/bash''' - tells linux this is a bash program and it should use a bash interpreter to execute it. |
| − | **'''#!/bin/bash''' - tells linux this is a bash program and it should use a bash interpreter to execute it. | + | ** '''#SBATCH''' - are SLURM parameters, for explanations of these please scroll down to SLURM Parameters Explanations section. |
| − | **'''# | + | ** '''sleep 20''' - Sleep 20 seconds (only used to simulate processing time for this example) |
| − | **'''sleep 20''' - Sleep 20 seconds (only used to simulate processing time for this example) | + | ** '''echo "Test Script"''' - Output some text to the screen when job completes ( simulate output for this example) |
| − | **'''echo "Test Script"''' - Output some text to the screen when job completes ( simulate output for this example) | + | * For example if you would like to run a blast job you may simply replace the last two line with the following |
| − | *For example if you would like to run a blast job you may simply replace the last two line with the following | + | |
| − | <pre>module load | + | <pre>module load BLAST |
blastall -p blastn -d nt -i input.fasta -e 10 -o output.result -v 10 -b 5 -a 5 | blastall -p blastn -d nt -i input.fasta -e 10 -o output.result -v 10 -b 5 -a 5 | ||
</pre> | </pre> | ||
| − | *Note: the module commands are explained under the '''Environment Modules''' section. | + | * Note: the module commands are explained under the '''Environment Modules''' section. |
| − | ==== | + | ====SLURM Parameters Explanations:==== |
| − | + | * To view all possible parameters | |
| + | ** Run '''man sbatch''' at the command line | ||
| + | ** Go to [https://slurm.schedmd.com/sbatch.html https://slurm.schedmd.com/sbatch.html] to view the man page online | ||
| − | {| | + | {| border="1" class='wikitable' |
|- | |- | ||
| − | ! Command | + | !|Command |
| − | ! Description | + | !|Description |
|- | |- | ||
| − | | # | + | ||#SBATCH -p PARTITION |
| − | | | + | ||Run the job on a specific queue/partition. This defaults to the "normal" queue |
|- | |- | ||
| − | | # | + | ||#SBATCH -D /tmp/working_dir |
| − | | Run the | + | ||Run the script from the /tmp/working_dir directory. This defaults to the current directory you are in. |
|- | |- | ||
| − | | # | + | ||#SBATCH -J ExampleJobName |
| − | | | + | ||Name of the job will be ExampleJobName |
|- | |- | ||
| − | | # | + | ||#SBATCH -e /path/to/errorfile |
| − | | | + | ||Split off the error stream to this file. By default output and error streams are placed in the same file. |
|- | |- | ||
| − | | # | + | ||#SBATCH -o /path/to/ouputfile |
| − | | | + | ||Split off the output stream to this file. By default output and error streams are placed in the same file. |
|- | |- | ||
| − | | # | + | ||#SBATCH --mail-user username@illinois.edu |
| − | | Send an e-mail to | + | ||Send an e-mail to specified email to receive job information. |
|- | |- | ||
| − | | # | + | ||#SBATCH --mail-type BEGIN, END, FAIL |
| − | | | + | ||Specifies when to send a message to email. You can select multiple of these with a comma separated list. Many other options exist. |
|- | |- | ||
| − | | # | + | ||#SBATCH -N X |
| − | | | + | ||Reserve X number of nodes. |
|- | |- | ||
| − | | # | + | ||#SBATCH -n X |
| − | | Reserve X number of | + | ||Reserve X number of cpus. |
|- | |- | ||
| − | | # | + | ||#SBATCH --mem=XG |
| − | | Reserve X gigabytes of RAM for the job. | + | ||Reserve X gigabytes of RAM for the job. |
|- | |- | ||
| − | | # | + | ||#SBATCH --gres=gpu:X |
| − | | | + | ||Reserve X NVIDIA GPUs. (Only on GPU queues) |
|} | |} | ||
| − | === Create a Job Array Script === | + | ===Create a Job Array Script=== |
| + | Making a new copy of the script and then submitting each one for every input data file is time consuming. An alternative is to make a job array using the '''#SBATCH --array''' option in your job script. The '''#SBATCH --array''' option allows many copies of the same script to be queued all at once. You can use the '''$SLURM_ARRAY_TASK_ID''' to differentiate between the different jobs in the array. A detailed example on how to do this is available at [[Job Arrays]] | ||
| − | + | ===Start An Interactive Session === | |
| − | + | * Use the '''srun''' commsnf if you would like to run a job interactively. | |
| − | + | <pre>srun --pty /bin/bash | |
| − | <pre> | ||
</pre> | </pre> | ||
| − | *This will automatically reserve you a slot on one of the compute nodes and will start a terminal session on it. | + | * This will automatically reserve you a slot on one of the compute nodes and will start a terminal session on it. |
| − | *Closing your terminal window will also kill your processes running in your interactive | + | * Closing your terminal window will also kill your processes running in your interactive srun session, therefore it's better to submit large jobs via non-interactive sbatch. |
| − | *To run an application with a user interface | + | |
| − | <pre> | + | === X11 Graphical Applications === |
| + | * To run an application with a user interface you will need to setup an Xserver on your computer [[Xserver Setup]] | ||
| + | *Then add the '''--x11''' parameter to your srun command | ||
| + | <pre> | ||
| + | srun --x11 --pty /bin/bash | ||
</pre> | </pre> | ||
| − | |||
| − | == View/Delete Submitted Jobs == | + | ==View/Delete Submitted Jobs== |
| + | ===Viewing Job Status=== | ||
| − | + | * To get a simple view of your current running jobs you may type: | |
| − | + | <pre>squeue -u userid | |
| − | <pre> | ||
</pre> | </pre> | ||
| − | *This command brings up a list of your current running jobs. | + | * This command brings up a list of your current running jobs. |
| − | *The first number represents the job's ID number. | + | * The first number represents the job's ID number. |
| − | *Jobs may have different status flags: | + | * Jobs may have different status flags: |
| − | **'''R''' = job is currently running | + | ** '''R''' = job is currently running |
| − | ** | + | |
| − | + | ||
| − | * | + | * For more detailed view type: |
| + | |||
| + | <pre>squeue -l</pre> | ||
| + | * This will return a list of all nodes, their slot availability, and your current jobs. | ||
| + | |||
| + | ===List Queues=== | ||
| + | |||
| + | * Simple view | ||
| − | + | <pre>sinfo</pre>This will show all queues as well as which nodes in those queues are fully used (alloc), partially used (mix), unused (idle), or unavailable (down). | |
| − | <pre> | ||
| − | |||
| − | |||
| − | * | + | ===List All Jobs on Cluster With Nodes=== |
| − | <pre> | + | <pre>squeue</pre> |
| + | ===Deleting Jobs=== | ||
| + | |||
| + | * Note: You can only delete jobs which are owned by you. | ||
| + | * To delete a job by job-ID number: | ||
| + | * You will need to use '''scancel''', for example to delete a job with ID number 5523 you would type: | ||
| + | |||
| + | <pre>scancel 5523 | ||
</pre> | </pre> | ||
| − | * | + | * Delete all of your jobs |
| − | + | ||
| − | + | <pre>scancel -u userid | |
| − | <pre> | ||
</pre> | </pre> | ||
| − | === | + | ===Troubleshooting job errors=== |
| − | <pre> | + | |
| + | * To view job errors in case job status shows | ||
| + | |||
| + | <pre>scontrol show job 23451 | ||
</pre> | </pre> | ||
| − | |||
| − | + | ==Applications== | |
| − | + | ===Application Lists=== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | === | ||
| − | * | + | * View a list of installed applications at [[Biocluster Applications]] |
| − | + | * List of currently installed applications from the commmand line, run '''module avail''' | |
| − | |||
| − | |||
| − | + | ===Application Installation=== | |
| − | |||
| − | == | ||
| − | + | * Please email '''help@igb.illinois.edu''' to request new application or version upgrades | |
| + | * The Biocluster uses EasyBuild to build and install software. You can read more about EasyBuild at [https://github.com/easybuilders/easybuild https://github.com/easybuilders/easybuild] | ||
| + | * The Biocluster EasyBuild scripts are located at [https://github.com/IGB-UIUC/easybuild https://github.com/IGB-UIUC/easybuild] | ||
| − | * | + | ===Environment Modules=== |
| − | * | + | * The Biocluster uses the Lmod modules package to manage the software that is installed. You can read more about Lmod at [https://lmod.readthedocs.io/en/latest/ https://lmod.readthedocs.io/en/latest/] |
| + | * To use an application, you need to use the '''module''' command to load the settings for an application | ||
| + | * To load a particular environment for example QIIME/1.9.1, simply run this command: | ||
| − | + | <pre>module load QIIME/1.9.1 | |
| + | </pre> | ||
| + | * If you would like to simply load the latest version, run the the command without the /1.9.1 (version number): | ||
| − | * | + | <pre>module load QIIME |
| + | </pre> | ||
| + | * To view which environments you have loaded simply run '''module list''': | ||
| − | + | <pre>bash-4.1$ module list | |
| + | Currently Loaded Modules: | ||
| + | 1) BLAST/2.2.26-Linux_x86_64 2) QIIME/1.9.1 | ||
| + | </pre> | ||
| + | * When submitting a job using a sbatch script you will have to add the '''module load qiime/1.5.0''' line before running qiime in the script. | ||
| + | * To unload a module simply run '''module unload''': | ||
| − | * | + | <pre>module unload QIIME |
| + | </pre> | ||
| + | * Unload all modules | ||
| + | <pre>module purge | ||
| + | </pre> | ||
| + | === Containers === | ||
| + | *The Biocluster cluster supports Singularity to run containers. | ||
| + | *The guide on how to use them is at [[Biocluster Singularity]] | ||
| − | * | + | === R Packages === |
| − | <pre> | + | *We have a local mirror of the [https://cran.r-project.org/ CRAN] and [https://www.bioconductor.org/ Bioconductor]. This allows you to install packages through an interactive session into your home folder. |
| + | *To install a package, run an interactive session | ||
| + | <pre> | ||
| + | srun --pty /bin/bash | ||
</pre> | </pre> | ||
| − | * | + | *Load the R module |
| − | <pre>module load | + | <pre> |
| + | module load R/4.4.0-IGB-gcc-8.2.0 | ||
</pre> | </pre> | ||
| − | * | + | *Run R |
| − | <pre> | + | <pre> |
| − | + | R | |
| − | + | </pre> | |
| + | *For CRAN packages, run install.packages() | ||
| + | <pre> | ||
| + | install.packages('shape'); | ||
| + | </pre> | ||
| + | *For Bioconductor packages, the BiocManager package is already installed. You just need to run BiocManager::install to install a package | ||
| + | <pre> | ||
| + | BiocManager::install('dada2') | ||
</pre> | </pre> | ||
| − | * | + | *If the package requires an external dependencies, you should email us to get it install centrally. |
| − | * | + | |
| − | <pre>module | + | === Python Packages === |
| + | * Most python packages in pypi, [https://pypi.org/ https://pypi.org/], are now precompiled. | ||
| + | * If the package is in pypi, from the biologin nodes, module load the Python version you want to use | ||
| + | <pre> | ||
| + | module load Python/3.10.1-IGB-gcc-8.2.0 | ||
</pre> | </pre> | ||
| − | * | + | * Then run pip install to install the package |
| − | <pre> | + | <pre> |
| + | pip install package_name | ||
</pre> | </pre> | ||
| − | == | + | * This might not work if it needs to compile the package or it needs to compile dependency packages. If that is the case, then email [mailto::help@igb.illinois.edu help@igb.illinois.edu] and we can get it installed. |
| + | === Jupyter === | ||
| + | * The biocluster has jupyterhub installed to allow you to run juypter notebooks at https://bioapps3.igb.illinois.edu/jupyter | ||
| + | * We have a guide on how to setup custom jupyter conda environments at [[Biocluster Jupyter]] | ||
| + | |||
| + | === Alphafold === | ||
| + | * The biocluster has Alphafold installed. There is specific instructions that need to be follow to run it on the biocluster. The guide is at [[Biocluster Alphafold]] | ||
| − | == | + | == Mirror Service - Genomic Databases == |
| + | * Biocluster provides mirrors of public accessible genomic databases | ||
| + | * If a database is not installed and it is publicly accessible, email [mailto::help@igb.illinois.edu help@igb.illinois.edu] and we can get it installed. | ||
| + | * If it is private database, then it must be placed in your home folder or a private group or lab folder. | ||
| + | * A list of databases is a located at [[Biocluster Mirrors]] | ||
| − | * | + | ==Transferring data files== |
| + | ===Transferring using SFTP/SCP=== | ||
| + | ====Using WinSCP==== | ||
| + | *Download WinSCP installation package from http://winscp.net/eng/download.php#download2 and install it. | ||
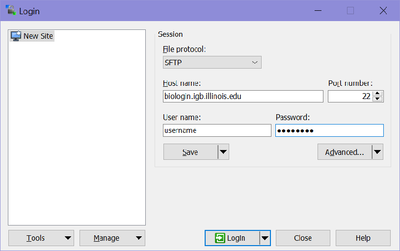
| + | *Once installed Run WinSCP >> enter biologin.igb.illinois.edu for the Host name >> Enter your IGB user name and password and click Login | ||
| + | [[File:Bioclustertransfer.png|400px]] | ||
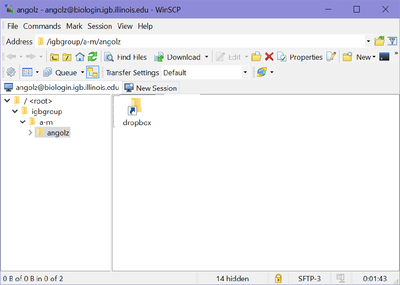
| + | *Once you hit "Login, you should be connected to your Biocluster home folder, as shown below. | ||
| + | [[File:Bioclustertransfer2.png|400px]] | ||
| + | *From here you should be able to download or transfer your files. | ||
| − | === | + | ====Using CyberDuck==== |
| − | * | + | *To download cyberduck go to [http://cyberduck.ch http://cyberduck.c] and click on the large Zip icon to download. |
| − | * | + | *Once cyberduck is installed on OSX you may start the program. |
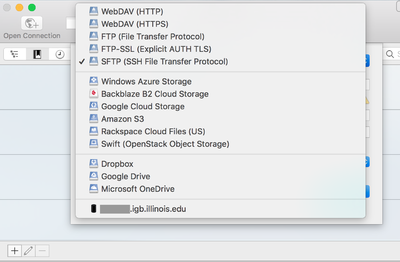
| − | + | *Click on '''Open Connection.''' | |
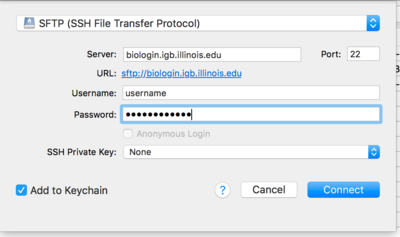
| + | *From the drop down menu at the top of the opopup window select '''SFTP(SSH File Transfer Protocol)''' | ||
| + | [[File:Cyberduck screenshot sftp.png|400px]] | ||
| − | + | *Now in the '''Server:''' input box enter '''biologin.igb.illinois.edu''' and for Username and password enter your IGB credentials. | |
| − | + | [[File:Cyberduckbiologin.png|400px]] | |
| − | * | + | |
| − | + | *Click '''Connect.''' | |
| − | + | *You may now download or transfer your files. | |
| − | + | *'''NOTICE:''' Cyberduck by default wants to open multiple connections for transferring files. The biocluster firewall limits you to 4 connections a minute. This can cause transfers to timeout. You can change Cyberduck to only use 1 connection by going to '''Preferences->Transfers->Transfer Files'''. Select '''Open Single Connection'''. | |
| − | |||
| − | + | ===Transferring using Globus=== | |
| − | * | + | * The biocluster has a Globus endpoint setup. The '''Collection Name''' is '''biocluster.igb.illinois.edu''' |
| − | + | * Globus allows the transferring of very large files reliably. | |
| − | + | * A guide on how to use Globus is [[Globus|here]] | |
| − | |||
| − | |||
| − | * | ||
| − | |||
| − | |||
| − | * | ||
| − | |||
| − | === | + | ===Core-Server=== |
| − | * | + | *The core-server is mounted on the biologin nodes at /private_stores/core-server. |
| − | *It can | + | *It is read-only; meaning you can only transfer data from the core-server to Biocluster. You cannot transfer any data from Biocluster to the core-server. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== References == | == References == | ||
| − | *[ | + | * OpenHPC [https://openhpc.community/ https://openhpc.community/] |
| + | * SLURM Job Scheduler Documentation - [https://slurm.schedmd.com/ https://slurm.schedmd.com/] | ||
| + | * Rosetta Stone of Schedulers - [https://slurm.schedmd.com/rosetta.pdf https://slurm.schedmd.com/rosetta.pdf] | ||
| + | * SLURM Quick Refernece - [https://slurm.schedmd.com/pdfs/summary.pdf https://slurm.schedmd.com/pdfs/summary.pdf] | ||
| + | * GPFS Filesystem [https://en.wikipedia.org/wiki/IBM_Spectrum_Scale https://en.wikipedia.org/wiki/IBM_Spectrum_Scale] | ||
| + | * Lmod Module Homepage - [https://www.tacc.utexas.edu/research-development/tacc-projects/lmod https://www.tacc.utexas.edu/research-development/tacc-projects/lmod] | ||
| + | * Lmod Documentation - [https://lmod.readthedocs.io/en/latest/ https://lmod.readthedocs.io/en/latest/] | ||
Latest revision as of 14:59, 22 August 2024
Contents
- 1 Quick Links
- 2 Description
- 3 Cluster Specifications
- 4 Storage
- 5 Calculate Usage (/private_stores)
- 6 Queue Costs
- 7 Gaining Access
- 8 Cluster Rules
- 9 How To Log Into The Cluster
- 10 How To Submit A Cluster Job
- 11 View/Delete Submitted Jobs
- 12 Applications
- 13 Mirror Service - Genomic Databases
- 14 Transferring data files
- 15 References
Quick Links[edit]
- Main Site - http://biocluster.igb.illinois.edu
- Request Account - http://www.igb.illinois.edu/content/biocluster-account-form
- Cluster Accounting - https://bioapps3.igb.illinois.edu/accounting/
- Cluster Monitoring - https://bioapps3.igb.illinois.edu/ganglia/
- SLURM Script Generator - http://www-app.igb.illinois.edu/tools/slurm/
- Biocluster Applications - https://help.igb.illinois.edu/Biocluster_Applications
- Biocluster Introduction Presentation - Intro_to_Biocluster_Spring_2022.pptx
Description[edit]
Biocluster is the High Performance Computing (HPC) resource for the Carl R Woese Institute for Genomic Biology (IGB) at the University of Illinois at Urbana-Champaign (UIUC). Containing 2824 cores and over 27.7 TB of RAM, Biocluster has a mix of various RAM and CPU configurations on nodes to best serve the various computation needs of the IGB and the Bioinformatics community at UIUC. For storage, Biocluster has 1.3 Petabytes of storage on its GPFS filesystem for reliable high speed data transfers within the cluster. Networking in Biocluster is either 1, 10 or 40 Gigibit ethernet depending on the class of node and its data transfer needs.
- The biocluster is not an authorized location to store HIPAA data.
- If you need to update the CFOP associated with your account, please send an email with the new CFOP to help@igb.illinois.edu.
Cluster Specifications[edit]
| Queue Name | Nodes | Cores (CPUs) per Node | Memory | Networking | Scratch Space /scatch | GPUs |
|---|---|---|---|---|---|---|
| normal (default) | 5 Supermicro SYS-2049U-TR4 | 72 Intel Xeon Gold 6150 @2.7 GHz | 1.2TB | 40GB Ethernet | 7TB SSD | |
| lowmem | 8 Supermicro SYS-F618R2-RTN+ | 12 Intel Xeon E5-2603 v4 @ 1.70Ghz | 64GB RAM | 10GB Ethernet | 192GB | |
| gpu | 1 Supermicro | 28 Intel Xeon E5-2680 @ 2.4Ghz | 256GB | 1GB Ethernet | 1TB SSD | 4 NVIDIA GeForce GTX 1080 Ti |
| classroom | 10 Dell Poweredge R620 | 24 Intel Xeon E5-2697 v2 @ 2.7Ghz | 384GB | 10GB Ethernet | 750GB |
Storage[edit]
Information[edit]
- The storage system is a GPFS filesystem with 1.3 Petabytes of total disk space with 2 copies of the data. This data is NOT backed up.
- The data is spread across 8 GPFS storage nodes.
Cost[edit]
On April 1, 2021, CNRG was informed by campus that we were required to start billing external users paying with a credit card an external rate. This rate was given to us by campus and is obtained by adding the 31.7% F&A rate and adding the standard 2.3% credit card fee. This external rate is only charged to users paying with a credit card.
| Internal Cost (Per Terabyte Per Month) | External Cost (Per Terabyte Per Month) |
|---|---|
| $8.75 | $11.73 |
Calculate Usage (/home)[edit]
- Each Month, you will receive a bill on your monthly usage. We take a snapspot of usage daily. Then we average out the 95 percentile of daily snapspots to get an average usage for the month.
- You can calculate your usage using the du command. An example is below. The result will be double what you are billed as their is 2 copies of the data. Make sure to divide by 2.
du -h /home/a-m/username
Calculate Usage (/private_stores)[edit]
- These are private data storage nodes. They do not get billed monthly.
- The filesystems are XFS shared over NFS.
- To calculate usage, use the du command
du -h /private_stores/shared/directory
Queue Costs[edit]
The cost for each job is dependent on which queue it is submitted to. Listed below are the different queues on the cluster with their cost. Although the service is billed by the second, the rates below are what it would cost per day to use a resource, so that it would be more easily understood. For standard computation, the CPU cost and the memory cost are compared and the highest is billed. For GPU bills the cost of the CPU or memory is added to that of the GPU.
Usage is charge by the second. The costs listed below are what it would cost per day. The CPU cost and memory cost are compared and the largest is what is billed.
| Queue Name | CPU Cost | External CPU Cost | Memory Cost | External Memory Cost | GPU Cost | External GPU Cost |
|---|---|---|---|---|---|---|
| normal (default) | $1.19 | $1.59 | $0.08 | $0.09 | NA | NA |
| lowmem | $0.50 | $0.67 | $0.10 | $0.13 | NA | NA |
| GPU | $2.00 | $2.68 | $0.44 | $0.59 | $2.00 | $2.68 |
On April 1, 2021, CNRG was informed by campus that we were required to start billing external users paying with a credit card an external rate. This rate was given to us by campus and is obtained by adding the 31.7% F&A rate and adding the standard 2.3% credit card fee. This external rate is only charged to users paying with a credit card.
Gaining Access[edit]
- Please fill out the form at http://www.igb.illinois.edu/content/biocluster-account-form to request access to the Biocluster.
Cluster Rules[edit]
- Running jobs on the head node or login nodes are strictly prohibited. Running jobs on the head node could cause the entire cluster to crash and affect everyone's jobs on the cluster. Any program found to be running on the headnode will be stopped immediately and your account could be locked. You can start an interactive session to login to a node to manual run programs.
- Installing Software Please email help@igb.illinois.edu for any software requests. Compiled software will be installed in /home/apps. If its a standard RedHat package (rpm), it will be installed in their default locations on the nodes.
- Creating or Moving over Programs: Programs you create or move to the cluster should be first tested by you outside the cluster for stability. Once your program is stable, then it can be moved over to the cluster for use. Unstable programs that cause problems with the cluster can result in your account being locked. Programs should only be added by CNRG personnel and not compiled in your home directory.
- Reserving Memory: SLURM allows the user to specify the amount of memory they want their program to use.. If your job tries to use more memory than you have reserved, the job will run out of memory and die. Make sure to specify the correct amount of memory.
- Reserving Nodes and Processors: For each job, you must reserve the correct number of nodes and processors. By default you are reserved 1 processor on 1 node. If you are running a multiple processor job or a MPI job you need to reserve the appropriate amount. If you do not reserve the correct amount, the cluster will confine your job to that limit, increasing its runtime.
How To Log Into The Cluster[edit]
- You will need to use an SSH client to connect.
- NOTICE The login hostname is biologin.igb.illinois.edu
On Windows[edit]
- You can download Putty from http://www.chiark.greenend.org.uk/~sgtatham/putty/download.html
- Install Putty and run it, in the Host Name input box enter biologin.igb.illinois.edu
- Hit Open and login using your IGB account credentials.
On Mac OS X[edit]
- Simply open the terminal under Go >> Utilities >> Terminal
- Type in ssh username@biologin.igb.illinois.edu where username is your NetID.
- Hit the Enter key and type in your IGB password.
How To Submit A Cluster Job[edit]
- The cluster runs the SLURM queuing and resource mangement program.
- All jobs are submitted to SLURM which distributes them automatically to the Nodes.
- You can find all of the parameters that SLURM uses at https://slurm.schedmd.com/quickstart.html
- You can use our SLURM Generation Utility to help you learn to generate job scripts http://www-app.igb.illinois.edu/tools/slurm/
Create a Job Script[edit]
- You must first create a SLURM job script file in order to tell SLURM how and what to execute on the nodes.
- Type the following into a text editor and save the file test.sh
#!/bin/bash #SBATCH -p normal #SBATCH --mem=1g #SBATCH -N 1 #SBATCH -n 1 sleep 20 echo "Test Script"
- You just created a simple SLURM Job Script.
- To submit the script to the cluster, you will use the sbatch command.
sbatch test.sh
- Line by line explanation
- #!/bin/bash - tells linux this is a bash program and it should use a bash interpreter to execute it.
- #SBATCH - are SLURM parameters, for explanations of these please scroll down to SLURM Parameters Explanations section.
- sleep 20 - Sleep 20 seconds (only used to simulate processing time for this example)
- echo "Test Script" - Output some text to the screen when job completes ( simulate output for this example)
- For example if you would like to run a blast job you may simply replace the last two line with the following
module load BLAST blastall -p blastn -d nt -i input.fasta -e 10 -o output.result -v 10 -b 5 -a 5
- Note: the module commands are explained under the Environment Modules section.
SLURM Parameters Explanations:[edit]
- To view all possible parameters
- Run man sbatch at the command line
- Go to https://slurm.schedmd.com/sbatch.html to view the man page online
| Command | Description |
|---|---|
| #SBATCH -p PARTITION | Run the job on a specific queue/partition. This defaults to the "normal" queue |
| #SBATCH -D /tmp/working_dir | Run the script from the /tmp/working_dir directory. This defaults to the current directory you are in. |
| #SBATCH -J ExampleJobName | Name of the job will be ExampleJobName |
| #SBATCH -e /path/to/errorfile | Split off the error stream to this file. By default output and error streams are placed in the same file. |
| #SBATCH -o /path/to/ouputfile | Split off the output stream to this file. By default output and error streams are placed in the same file. |
| #SBATCH --mail-user username@illinois.edu | Send an e-mail to specified email to receive job information. |
| #SBATCH --mail-type BEGIN, END, FAIL | Specifies when to send a message to email. You can select multiple of these with a comma separated list. Many other options exist. |
| #SBATCH -N X | Reserve X number of nodes. |
| #SBATCH -n X | Reserve X number of cpus. |
| #SBATCH --mem=XG | Reserve X gigabytes of RAM for the job. |
| #SBATCH --gres=gpu:X | Reserve X NVIDIA GPUs. (Only on GPU queues) |
Create a Job Array Script[edit]
Making a new copy of the script and then submitting each one for every input data file is time consuming. An alternative is to make a job array using the #SBATCH --array option in your job script. The #SBATCH --array option allows many copies of the same script to be queued all at once. You can use the $SLURM_ARRAY_TASK_ID to differentiate between the different jobs in the array. A detailed example on how to do this is available at Job Arrays
Start An Interactive Session[edit]
- Use the srun commsnf if you would like to run a job interactively.
srun --pty /bin/bash
- This will automatically reserve you a slot on one of the compute nodes and will start a terminal session on it.
- Closing your terminal window will also kill your processes running in your interactive srun session, therefore it's better to submit large jobs via non-interactive sbatch.
X11 Graphical Applications[edit]
- To run an application with a user interface you will need to setup an Xserver on your computer Xserver Setup
- Then add the --x11 parameter to your srun command
srun --x11 --pty /bin/bash
View/Delete Submitted Jobs[edit]
Viewing Job Status[edit]
- To get a simple view of your current running jobs you may type:
squeue -u userid
- This command brings up a list of your current running jobs.
- The first number represents the job's ID number.
- Jobs may have different status flags:
- R = job is currently running
- For more detailed view type:
squeue -l
- This will return a list of all nodes, their slot availability, and your current jobs.
List Queues[edit]
- Simple view
sinfo
This will show all queues as well as which nodes in those queues are fully used (alloc), partially used (mix), unused (idle), or unavailable (down).
List All Jobs on Cluster With Nodes[edit]
squeue
Deleting Jobs[edit]
- Note: You can only delete jobs which are owned by you.
- To delete a job by job-ID number:
- You will need to use scancel, for example to delete a job with ID number 5523 you would type:
scancel 5523
- Delete all of your jobs
scancel -u userid
Troubleshooting job errors[edit]
- To view job errors in case job status shows
scontrol show job 23451
Applications[edit]
Application Lists[edit]
- View a list of installed applications at Biocluster Applications
- List of currently installed applications from the commmand line, run module avail
Application Installation[edit]
- Please email help@igb.illinois.edu to request new application or version upgrades
- The Biocluster uses EasyBuild to build and install software. You can read more about EasyBuild at https://github.com/easybuilders/easybuild
- The Biocluster EasyBuild scripts are located at https://github.com/IGB-UIUC/easybuild
Environment Modules[edit]
- The Biocluster uses the Lmod modules package to manage the software that is installed. You can read more about Lmod at https://lmod.readthedocs.io/en/latest/
- To use an application, you need to use the module command to load the settings for an application
- To load a particular environment for example QIIME/1.9.1, simply run this command:
module load QIIME/1.9.1
- If you would like to simply load the latest version, run the the command without the /1.9.1 (version number):
module load QIIME
- To view which environments you have loaded simply run module list:
bash-4.1$ module list Currently Loaded Modules: 1) BLAST/2.2.26-Linux_x86_64 2) QIIME/1.9.1
- When submitting a job using a sbatch script you will have to add the module load qiime/1.5.0 line before running qiime in the script.
- To unload a module simply run module unload:
module unload QIIME
- Unload all modules
module purge
Containers[edit]
- The Biocluster cluster supports Singularity to run containers.
- The guide on how to use them is at Biocluster Singularity
R Packages[edit]
- We have a local mirror of the CRAN and Bioconductor. This allows you to install packages through an interactive session into your home folder.
- To install a package, run an interactive session
srun --pty /bin/bash
- Load the R module
module load R/4.4.0-IGB-gcc-8.2.0
- Run R
R
- For CRAN packages, run install.packages()
install.packages('shape');
- For Bioconductor packages, the BiocManager package is already installed. You just need to run BiocManager::install to install a package
BiocManager::install('dada2')
- If the package requires an external dependencies, you should email us to get it install centrally.
Python Packages[edit]
- Most python packages in pypi, https://pypi.org/, are now precompiled.
- If the package is in pypi, from the biologin nodes, module load the Python version you want to use
module load Python/3.10.1-IGB-gcc-8.2.0
- Then run pip install to install the package
pip install package_name
- This might not work if it needs to compile the package or it needs to compile dependency packages. If that is the case, then email help@igb.illinois.edu and we can get it installed.
Jupyter[edit]
- The biocluster has jupyterhub installed to allow you to run juypter notebooks at https://bioapps3.igb.illinois.edu/jupyter
- We have a guide on how to setup custom jupyter conda environments at Biocluster Jupyter
Alphafold[edit]
- The biocluster has Alphafold installed. There is specific instructions that need to be follow to run it on the biocluster. The guide is at Biocluster Alphafold
Mirror Service - Genomic Databases[edit]
- Biocluster provides mirrors of public accessible genomic databases
- If a database is not installed and it is publicly accessible, email help@igb.illinois.edu and we can get it installed.
- If it is private database, then it must be placed in your home folder or a private group or lab folder.
- A list of databases is a located at Biocluster Mirrors
Transferring data files[edit]
Transferring using SFTP/SCP[edit]
Using WinSCP[edit]
- Download WinSCP installation package from http://winscp.net/eng/download.php#download2 and install it.
- Once installed Run WinSCP >> enter biologin.igb.illinois.edu for the Host name >> Enter your IGB user name and password and click Login
- Once you hit "Login, you should be connected to your Biocluster home folder, as shown below.
- From here you should be able to download or transfer your files.
Using CyberDuck[edit]
- To download cyberduck go to http://cyberduck.c and click on the large Zip icon to download.
- Once cyberduck is installed on OSX you may start the program.
- Click on Open Connection.
- From the drop down menu at the top of the opopup window select SFTP(SSH File Transfer Protocol)
- Now in the Server: input box enter biologin.igb.illinois.edu and for Username and password enter your IGB credentials.
- Click Connect.
- You may now download or transfer your files.
- NOTICE: Cyberduck by default wants to open multiple connections for transferring files. The biocluster firewall limits you to 4 connections a minute. This can cause transfers to timeout. You can change Cyberduck to only use 1 connection by going to Preferences->Transfers->Transfer Files. Select Open Single Connection.
Transferring using Globus[edit]
- The biocluster has a Globus endpoint setup. The Collection Name is biocluster.igb.illinois.edu
- Globus allows the transferring of very large files reliably.
- A guide on how to use Globus is here
Core-Server[edit]
- The core-server is mounted on the biologin nodes at /private_stores/core-server.
- It is read-only; meaning you can only transfer data from the core-server to Biocluster. You cannot transfer any data from Biocluster to the core-server.
References[edit]
- OpenHPC https://openhpc.community/
- SLURM Job Scheduler Documentation - https://slurm.schedmd.com/
- Rosetta Stone of Schedulers - https://slurm.schedmd.com/rosetta.pdf
- SLURM Quick Refernece - https://slurm.schedmd.com/pdfs/summary.pdf
- GPFS Filesystem https://en.wikipedia.org/wiki/IBM_Spectrum_Scale
- Lmod Module Homepage - https://www.tacc.utexas.edu/research-development/tacc-projects/lmod
- Lmod Documentation - https://lmod.readthedocs.io/en/latest/