Biocluster2
Quick Links
- Main Site - http://biocluster2.igb.illinois.edu
- Request Account - http://www.igb.illinois.edu/content/biocluster-account-form
- Cluster Accounting - https://biocluster2.igb.illinois.edu/accounting/
- Cluster Monitoring - http://biocluster2.igb.illinois.edu/ganglia/
- SLURM Script Generator - http://www-app2.igb.illinois.edu/tools/slurm/
Description
Biocluster is the High Performance Computing (HPC) resource for the Carl R Wose Institute for Genomic Biology (IGB) at the University of Illinois at Urbana-Champaign (UIUC). Containing 2824 cores and over 27.7 TB of RAM, Biocluster has a mix of various RAM and CPU configurations on nodes to best serve the various computation needs of the IGB and the Bioinformatics community at UIUC. For storage, Biocluster has 700TB of storage on its GPFS filesystem for reliable high speed data transfers within the cluster. Networking in Biocluster is either 1 or 10 Gigibit ethernet depending on the class of node and its data transfer needs.
Cluster Specifications
Default Queue and Highthroughput Queue
- 20 Dell PowerEdge R620 Servers
- 24 Intel Xeon E5-2697 @ 2.7GHz CPU Cores per node
- 384 Gigabytes of RAM per Node
Budget Queue
- 10 Dell Poweredge R410
- 8 Intel Xeon E5530 @ 2.4Ghz CPU Cores per node
- 32 or 48 Gigabytes of RAM per Node
Large Memory Queue
- 1 Node - Dell R910
- 24 Intel Xeon E7540 @ 2.0GHz CPUs per node
- 1024 Gigabytes (1TB) of RAM
Blacklight Queue
- 4 SGI UV1000 Nodes
- 384 Intel Xeon X7542 @ 2.67 CPUs per node
- 2 TB of Ram per node
Storage
Information
The storage system is a GPFS filesystem with 650 Terabytes of disk space. This data is NOT backed up.
Cost
| Cost (Per Terabyte Per Month) |
|---|
| $10 |
Queue Costs
The cost for each job is dependent on which queue it is submitted to. Listed below are the different queues on the cluster with their cost.
Usage is charge by the second. The CPU cost and memory cost are compared and the largest is what is billed.
| Queue Name | CPU Cost ($ per CPU per day) | Memory Cost ($ per GB per day) |
|---|---|---|
| default | $1.00 | $0.50 |
| highthroughput | $1.00 | $0.50 |
| largememory | $8.50 | $0.20 |
| blacklight |
$1.10 | $0.25 |
| budget |
$0.50 | $0.25 |
How To Get Biocluster
- Please fill out the form at http://www.igb.illinois.edu/content/biocluster-account-form to request access to the Biocluster.
Cluster Rules
- Running jobs on the head node are strictly prohibited. Running jobs on the head node could cause the entire cluster to crash and affect everyone's jobs on the cluster. Any program found to be running on the headnode will be stopped immediately and your account could be locked. You can start an interactive session to login to a node to manual run programs.
- Installing Software Please email help@igb.illinois.edu for any software requests. Compiled software will be installed in /home/apps. If its a standard RedHat package (rpm), it will be installed in their default locations on the nodes.
- Creating or Moving over Programs: Programs you create or move to the cluster should be first tested by you outside the cluster for stability. Once your program is stable, then it can be moved over to the cluster for use. Unstable programs that cause problems with the cluster can result in your account being locked. Programs should only be added by CNRG personnel and not compiled in your home directory.
- Reserving Memory: TORQUE allows the user to specify the amount of memory they want their program to use.. If your job tries to use more memory than you have reserved, the job will automatically get shutdown. Make sure to specify the correct amount of memory.
- Reserving Nodes and Processors: For each job, you must reserve the correct number of nodes and processors. By default you are reserved 1 processor on 1 node. If you are running a multiple processor job or a MPI job you need to reserve the appropriate amount. If you do not reserve the correct amount, the cluster will shutdown your job. Failure to comply with this policy will result in your account being locked.
How To Log Into The Cluster
- You will need to use an SSH client to connect.
On Windows
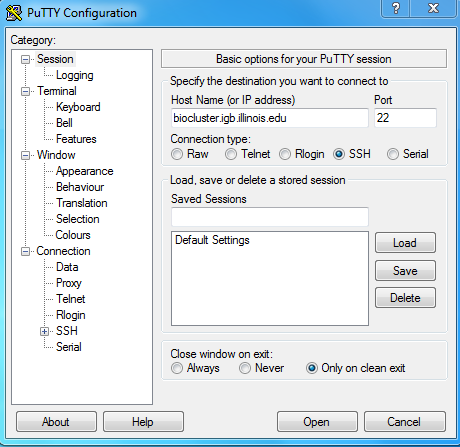
- You can download Putty from http://www.chiark.greenend.org.uk/~sgtatham/putty/download.html
- Install Putty and run it, in the Host Name input box enter biocluster.igb.illinois.edu
- Hit Open and login using your IGB account credentials.
On Mac OS X
- Simply open the terminal under Go >> Utilities >> Terminal
- Type in ssh username@biocluster.igb.illinois.edu where username is your NetID.
- Hit the Enter key and type in your IGB password.
How To Submit A Cluster Job
- The cluster runs the TORQUE queuing and resource mangement program.
- All jobs are submitted to TORQUE which distributes them automatically to the Nodes.
- You can find all of the parameters that TORQUE uses at http://www.adaptivecomputing.com/wp-content/media/pdf/Handout_TorqueTutorial_qsub.pdf
- You can use our Qsub Generation Utility to help you learn to generate job scripts http://www-app2.igb.illinois.edu/tools/qsub/
Create a Job Script
- You must first create a TORQUE job script file in order to tell TORQUE how and what to execute on the nodes.
- Type the following into a text editor and save the file test.sh ( Linux text editing )
#!/bin/bash #PBS -j oe sleep 20 echo "Test Script"
- Change the permissions on the script to allow execution.
chmod 775 test.sh
- You just created a simple PBS TORQUE Job Script.
- To submit the script to the cluster, you will use the qsub command.
qsub test.sh
- Line by line explanation
- #!/bin/bash - tells linux this is a bash program and it should use a bash interpreter to execute it.
- #PBS - are PBS parameters, for explanations of these please scroll down to PBS Parameters Explanations section.
- sleep 20 - Sleep 20 seconds (only used to simulate processing time for this example)
- echo "Test Script" - Output some text to the screen when job completes ( simulate output for this example)
- For example if you would like to run a blast job you may simply replace the last two line with the following
module load blast blastall -p blastn -d nt -i input.fasta -e 10 -o output.result -v 10 -b 5 -a 5
- Note: the module commands are explained under the Environment Modules section.
TORQUE PBS Parameters Explanations:
These are just a few parameter options, for more type man qsub while logged into the cluster.
| Command | Description |
|---|---|
| #PBS -S /bin/bash | The program will be using bash for its interpreter. Required. |
| #PBS -q QUEUENAME | Run the job on a specific queue. This defaults to the "default" queue |
| #PBS -d /tmp/working_dir | Run the script from the /tmp/working_dir directory. This defaults to your home directory (/home/a-m/username, /home/n-z/username). |
| #PBS -N ExampleJobName | Name of the job will be ExampleJobName |
| #PBS -j oe | Join the standard out and standard error streams together into one file. This file will be created in the working directory and will be named in this case test.sh.o# where # is the job number assigned by Torque. |
| #PBS -M username@illinois.edu | Send an e-mail to username@igb.illinois.edu to receive job information. |
| #PBS -m abe | Send email when job aborts, job begins, and job ends |
| #PBS -l | Reserves resources such as number of processors, memory, wallclock. |
| #PBS -l nodes=X:ppn=Y | Reserve X number of nodes with Y processors per node. |
| #PBS -l mem=XGB | Reserve X gigabytes of RAM for the job. |
| #PBS -l nodes=1:ppn=5,mem=4GB | To combine multiple resources, separate them with a comma |
Create a Job Array Script
Making a new copy of the script and then submitting each one for every input data file is time consuming. An alternative is to make a job array using the -t option in your submission script. The -t option allows many copies of the same script to be queued all at once. You can use the PBS_ARRAYID to differentiate between the different jobs in the array. A brief example on how to do this is available at Job Array Example
Start An Interactive Login Session On A Compute Node
- Use interactive qsub if you would like to run a job interactively such as running a quick perl script or run a quick test interactively on your data.
qsub -I
- This will automatically reserve you a slot on one of the compute nodes and will start a terminal session on it.
- Closing your terminal window will also kill your processes running in your interactive qsub session, therefor it's better to submit large jobs via non-interactive qsub.
- To run an application with a user interface run
qsub -I -X
- For this to work you will need to setup xserver on your computer Xserver Setup
View/Delete Submitted Jobs
Viewing Job Status
- To get a simple view of your current running jobs you may type:
qstat
- This command brings up a list of your current running jobs.
- The first number represents the job's ID number.
- Jobs may have different status flags:
- R = job is currently running
- W = job is waiting to be submitted (this may take a few seconds even when there are slots available so be patient)
- Eqw = There was an error running the job.
- S = Job is suspended (job overused the resources subscribed to it in the qsub command)
- For more detailed view type:
qstat -f
- This will return a list of all nodes, their slot availability, and your current jobs.
List Queues
- Simple view
qstat -q
- Advance view (Ex: where queue name is budget)
- max_user_queuable - max number of jobs allowed to be in the queue
- max_user_run - max number of jobs the queue will run at same time
qstat -Qf budget
List All Jobs on Cluster With Nodes
qstat -a -n
Deleting Jobs
- Note: You can only delete jobs which are owned by you.
- To delete a job by job-ID number:
- You will need to use qdel, for example to delete a job with ID number 5523 you would type:
qdel 5523
- Delete all of your jobs
qdel all
Troubleshooting job errors
- To view job errors in case job status shows Eqw or any other error in the status column use qstat -j, for example if job # 23451 failed you would type:
qstat -j 23451
View Job Details
- To view job details, you can use the qstat -f JOB_NUMBER, checkjob JOBNUMBER, tracejob JOBNUMBER to monitor your job.
qstat -f 12345
checkjob 12345
tracejob 12345
Applications
Application Lists
- List of currently installed applications, please go to Biocluster Applications
- List of currently installed applications from the commmand line, run module avail
Application Installation
- Please email help@igb.illinois.edu to request new application or version upgrades
Environment Modules
- To use an application, you need to use the module command to load the settings for an application
- To load a particular environment for example qiime/1.5.0, simply run this command:
module load qiime/1/5.0
- If you would like to simply load the latest version, run the the command without the /1.50 (version number):
module load qiime
- To view which environments you have loaded simply run module list:
bash-4.1$ module list Currently Loaded Modulefiles: 1) qiime
- When submitting a job using a qsub script you will have to add the module load qiime/1.5.0 line before running qiime in the script.
- To unload a module simply run module unload:
module unload qiime
- Unload all modules
module purge
Transferring data files
Transferring using SFTP/SCP
- In order to transfer files to the cluster from a personal Desktop/Laptop you may use WinSCP the same way you would use it to transfer files to the File Server.
Transferring using Globus
- The biocluster has a Globus endpoint setup. Then end point name is igb#biocluster.igb.illinois.edu
- Globus allows the transferring of very large files reliably.
- A guide on how to use Globus is here
Transferring from file server (Very Fast)
- To transfer files to the cluster form the file-server you will need to first setup Xserver on your machine. Please follow this guide to do so Xserver Setup.
- Once Xserver is setup on your personal machine you will need to SSH into the cluster using putty as mentioned above.
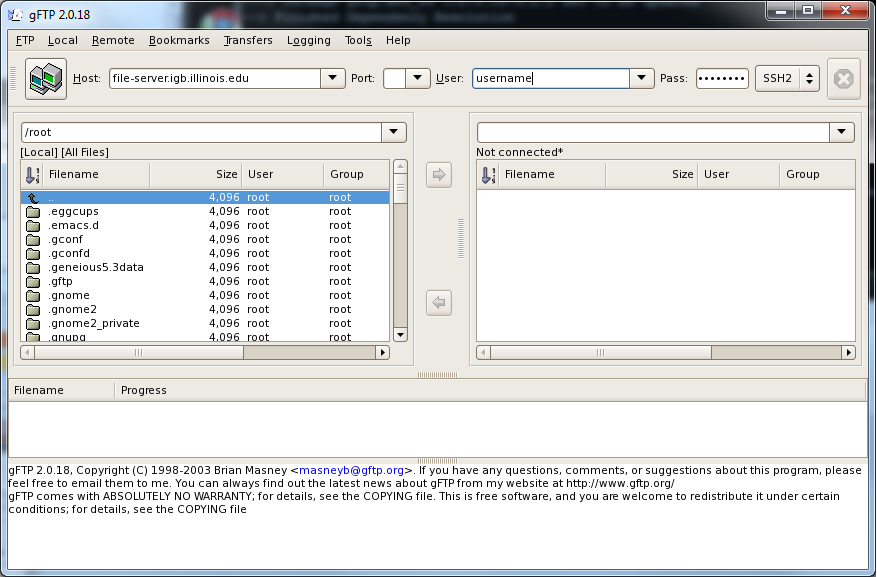
- Then start gFTP by typing in the terminal:
gftp
- This will launch a graphical interface for gFTP on your computer, it should look like this
- Enter the following into the gFTP user interface:
- Host: file-server.igb.illinois.edu
- Port: leave this box blank
- User: Your IGB username
- Pass: Your IGB password
- Select SSH2 from the drop down menu
- Hit enter and you should now be connected to the file-server from the cluster.
- You may now select files and folders from your home directories and click the arrows pointing in each direction to transfer files both ways.
- Please move files back to the file-server or your personal machine once you are done working with them on the cluster in order to allow others to use the space on the cluster for their jobs.
- Note: You may also use the standard command line tool "sftp" to transfer files if you do not want to use gFTP.
Transferring from Biotech FTP Server
- One option to transfer data from the Biotech FTP server is to use a program called wget.
- It can download 1 file or an entire directory.
- Replace USERNAME with the username provided by the Biotech Center.
The below example will download the file test_file.tar.gz
wget --user=USERNAME --ask-password ftp://ftp.biotech.illinois.edu/test_file.tar.gz
- If you want to download an entire directory, you need to have the -r parameter set. This will recursively go through an entire directory and download it.
wget -r --user=USERNAME --ask-password ftp://ftp.biotech.illinois.edu/test_dir/