Biocluster: Difference between revisions
Jump to navigation
Jump to search
| Line 168: | Line 168: | ||
*The first number represents the job's ID number. | *The first number represents the job's ID number. | ||
*Jobs may have different status flags: | *Jobs may have different status flags: | ||
**''' | **'''R''' = job is currently running | ||
**''' | **'''W''' = job is waiting to be submitted (this may take a few seconds even when there are slots available so be patient) | ||
**'''Eqw''' = There was an error running the job. | **'''Eqw''' = There was an error running the job. | ||
Revision as of 15:47, 18 November 2011
Biocluster
Cluster Specifications
The biocluster is a combination of our former clusters, which allows for a smoother operating environment. Users will decide which "queue" they will submit to instead of logging into a specific cluster.
Classroom Queue
- 25 Nodes
- Dell
- X X GHz Intel CPU's per Node
- XX Gigabytes of RAM per Node
Computation Queue
- 11 Nodes
- Dell R410 Servers
- 8 2.6 GHz Intel CPUs per Node
- 24 Gigabytes of RAM per Node
Large Memory Queue
- 2 Nodes
- Node 1 - Dell R900
- 16 2.4 GHz Intel CPUs
- 256 Gigabytes of RAM
- Node 2 - Dell R910
- 24 2.0 GHz Intel CPUs
- 1024 Gigabytes (1TB) of RAM
Cluster Status
- Resources Status
- Job Queue
- Cluster Accounting - View your cluster usage/charges
Usage Cost
Classroom Queue
- Currently used for TORQUE training purposes.
Computation Queue
- The daily cost (24-hour period) of computing on this cluster is about $1.37 per day per cpu.
Large Memory Queue
- The daily cost (24-hour period) of computing on this cluster is about $11.42 per day per cpu.
How To Get Cluster Access
IGB Affliated
- If you are a member of IGB you may send an email to help@igb.uiuc.edu requesting access to the cluster and CFOP account to charge cluster use to.
- You must also request your professor or theme leader to e-mail us at help@igb.uiuc.edu approving your access, unless you are a fellow.
Non-IGB Affliated
- You must have a note from someone affiliated with the IGB approving your use of the cluster.
- Once you are authorized to use the cluster you may go to room 2626 IGB in order to sign up for an IGB account
- Bring the approval letter and a CFOP account to charge cluster use, to room 2626 and our staff will provide you with access.
Cluster Rules
- Running jobs on the head node are strictly prohibited. If you would like to start an interactive session to run a quick script or test code please use qlogin. (Running jobs on the head node could cause it to crash and kill everyone's jobs on the slaves)
- Adding Packages: Packages (rpms) requested to be added are normally reviewed for a short while, and then installed on the slave and/or head nodes. Please be sure to specify which type of node it is needed on. Requests should be sent to help@igb.uiuc.edu.
- Creating or Moving over Programs: Programs you create or move to the cluster should be first tested by you outside the cluster for stability. Once you see your program is stable, it is safe to move them over for use in TORQUE. Unstable programs that cause problems with the cluster can cause your account to be locked in rare situations. Programs should only be added by CNRG personnel and not compiled in your home directory.
- Selecting which queue to run your job in: The biocluster, while being one big cluster, will have multiple queues which each have different hardware specifications. For example if you wanted to submit a qsub script called temp.sh to the large memory que you would execute the following command:
qusub -q largememory temp.sh
- Slots and Process Forking: For each process you create in a program you run through TORQUE, you must reserve a slot in the TORQUE. Normally this is done automatically (one slot for the one process you are submitting), but if you are spawning (also called forking) other processes from that process you submitted, you must reserve one slot per process created. Normally this is done as a serial process rather than a parallel process. For example if you would like to reserve 8 or more processing slots for a qsub script called temp.sh in the computation queue you would execute the following command:
qsub -l node=1:ppn=8 -q computation temp.sh
If you are doing this serially and create (fork/spawn) more than 8 processes, you only need to request 8 processes at a time. Failure to comply with this policy will result in your account being locked.
How To Log Into The Cluster
- You will need to use an SSH client to connect.
On Windows
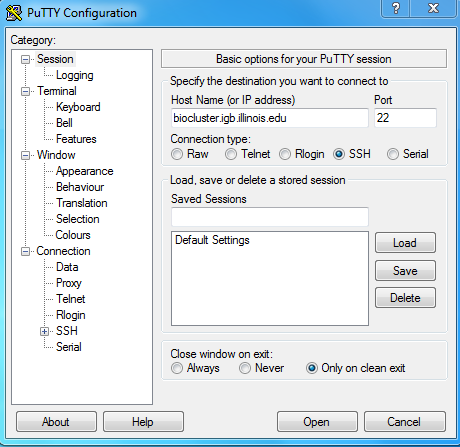
- You can download Putty from http://www.chiark.greenend.org.uk/~sgtatham/putty/download.html
- Install Putty and run it, in the Host Name input box enter biocluster.igb.illinois.edu
- Hit Open and login using your IGB account credentials.
On Mac OS X
- Simply open the terminal under Go >> Utilities >> Terminal
- Type in ssh YourUserName@biocluster.igb.illinois.edu where YourUserName is your IGB user name (NetID).
- Hit the Enter key and type in your IGB password.
How To Submit A Cluster Job
- The cluster runs TORQUE queuing and resource mangement program.
- All jobs are submitted to TORQUE which distributes them automatically to the Nodes.
Create a Job Script
- You must first create a TORQUE job script file in order to tell TORQUE how and what to execute on the nodes.
- This example demonstrates script creation using the vi text editor.
- First we need to create a file for the script
- In the command line type vi test.sh
- Now click i in order to enable text insertion.
- Type this example bash script into vi
#!/bin/bash #$ -j y #$ -S /bin/bash sleep 20 echo "Starting Blast Job"
- Click Esc to stop inserting text, type :w and click Enter to save the file.
- Type :q and click Enter to quit vi.
- You just created a simple SGE Job Script.
- The first line tells linux this is a bash program (#!/bin/bash) and it should use a bash interpreter to execute it.
- All lines with #$ are SGE parameters, for explanations of these please scroll down to SGE Parameters Explanations section.
- The last three lines are simple linux commands that will execute on the node in consecutive order.
- First sleep 20 tells linux to sleep the bash script for 20 seconds.
- Then echo "Starting Blast Job" will print to the output stream file Starting Blast Job.
- If you would like to run a blast job for example you may add this line at the end of the file.
/opt/Bio/ncbi/bin/blastall -p blastn -d nt -i input.fasta -e 10 -o output.result -v 10 -b 5 -a 5
TORQUE Parameters Explanations:
- These are just a few parameter options, for more type man qsub while logged into the cluster.
- #$ -cwd tells SGE to run the script from the current directory. This defaults to your home directory (/share/home/username). This could be problematic if the path on the head node is different than the path on the slave node.
- #$ -j y parameter tells SGE to join the errors and output streams together into one file. This file will be created in the directory described by #$ -cwd and will be named in this case test.sh.o# where # is the job number assigned by the SGE.
- #$ -S /bin/bash parameter tells the SGE that the program will be using bash for its interpreter. Required.
- #$ -N testJob3 parameter tells the SGE to name the job testJob3.
- #$ -M username@igb.illlinois.edu parameter tells the SGE to send an e-mail to username@igb.illinois.edu when the job is done.
- #$ -m abe parameter tells the SGE to send an e-mail to the e-mail defined using the above -M parameter when a job is aborted,begins or ends.
- #$ -pe serial 5 parameter tells the SGE to reserve 5 processors on a node for this job. This is only allowed if your program can actually use multiple processors natively, otherwise this is considered cluster abuse (In the example script test.sh, blastall has the parameter -a 5 which tells blast to run using 5 processors in this case telling SGE to reserve 5 processors is justified!)
- #$ -l h_data=1024M parameter tells the SGE to reserve 1024 Megabytes of RAM per core for this job.'
- #$ -q all.q@compute-0-5.local parameter tells the SGE to run the job on compute node compute-0-5.
- #$ -v PYTHONPATH=/opt/rocks/lib/python2.4/site-packages parameter tells SGE to set the PYTHONPATH environmental variable to /opt/rocks/lib/python2.4 for this job, this will work for any environmental variable not just PYTHONPATH
[1]
Submit Serial Job
- To submit the serial job you will use the qsub program. For example to submit test.sh TORQUE Job you would type:
qsub test.sh
- You may also define the TORQUE parameters from the section above as qsub parameters instead of defining them in the script file. Example:
qsub -j y -S /bin/bash test.sh
Submit Parallel Job
- To submit the parallel job you will use the qsub program.
- For more information please refer to this page Submitting TORQUE Jobs
- To distribute the jobs evenly across the reserved nodes you will have to to use orte_rr instead of the orte parallel environment. This will distribute the MPI jobs to each node in round robin order. While the default orte will make sure to fill all slots on the node before moving to the next one.
Start An Interactive Login Session On A Compute Node
- Use qlogin if you would like to run a job interactively such as running a quick perl script or run a quick test interactively on your data.
qlogin
- This will automatically reserve you a slot on one of the compute nodes and will start a terminal session on it.
- Closing your terminal window will also kill your processes running in your qlogin session, therefor it's better to submit large jobs via qsub.
View/Delete Submitted Jobs
Viewing Job Status
- To get a simple view of your current running jobs you may type:
qstat
- This command brings up a list of your current running jobs.
- The first number represents the job's ID number.
- Jobs may have different status flags:
- R = job is currently running
- W = job is waiting to be submitted (this may take a few seconds even when there are slots available so be patient)
- Eqw = There was an error running the job.
- For more detailed view type:
qstat -f
- This will return a list of all nodes, their slot availability, and your current jobs.
Deleting Jobs
- Note: You can only delete jobs which are owned by you.
- To delete a job by job-ID number:
- You will need to use qdel, for example to delete a job with ID number 5523 you would type:
qdel 5523
Troubleshooting job errors
- To view job errors in case job status shows Eqw or any other error in the status column use qstat -j, for example if job # 23451 failed you would type:
qstat -j 23451
Installed Applications
- To list available applications while logged into the head node you may run:
ls /share/apps/
- The applications path on the compute nodes is also:
/share/apps/
- Notice: As mentioned in the rules you may not run these applications on the head node (for more details please see rules at the top of the page).
Compute node paths
- Installed programs folder path:
/share/apps/
- Add java binary path to qsub script:
export PATH=$PATH:/usr/java/latest/bin
- Add Python 2.4 modules path to qsub script:
export PYTHONPATH=$PYTHONPATH:/opt/rocks/lib/python2.4/site-packages
- NCBI blast binary path:
/opt/Bio/ncbi/bin/blastall
Transferring data files
Transferring from personal machine
- In order to transfer files to the cluster from a personal Desktop/Laptop you may use WinSCP the same way you would use it to transfer files to the File Server.
Transferring from file server (Very Fast)
- To transfer files to the cluster form the file-server you will need to first setup Xserver on your machine. Please follow this guide to do so Xserver Setup.
- Once Xserver is setup on your personal machine you will need to SSH into the cluster using putty as mentioned above.
- Then start gFTP by typing in the terminal:
gftp
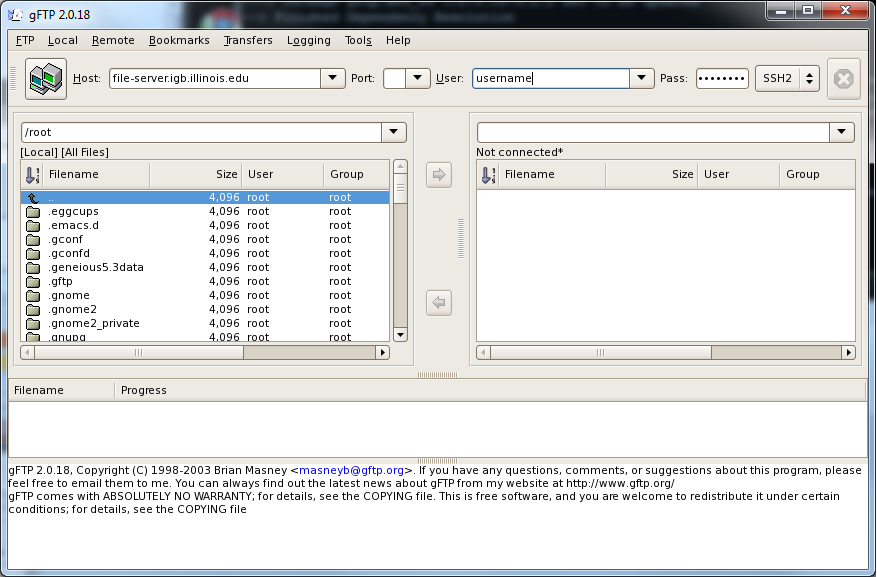
- This will launch a graphical interface for gFTP on your computer, it should look like this
- Enter the following into the gFTP user interface:
- Host: file-server.igb.uiuc.edu
- Port: leave this box blank
- User: Your IGB username
- Pass: Your IGB password
- Select SSH2 from the drop down menu
- Hit enter and you should now be connected to the file-server from the cluster.
- You may now select files and folders from your home directories and click the arrows pointing in each direction to transfer files both ways.
- Please move files back to the file-server or your personal machine once you are done working with them on the cluster in order to allow others to use the space on the cluster for their jobs.
- Note: You may also use the standard command line tool "sftp" to transfer files if you do not want to use gFTP.
Disk Usage
- Currently there are no disk usage specifications.
- If there is a special interest or recommendations please let us know at help@igb.illinois.edu