Difference between revisions of "Using Bioarchive"
(→Data transfer from a computer) |
|||
| (96 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | = | + | [[File:T950.jpeg|right|border|400px]] |
| − | + | __TOC__ | |
| + | =Bioarchive= | ||
| + | Bioarchive is set up for long-term data storage at a low upfront cost, which allows grant PIs to preserve data well beyond the funding period. Storage space can be purchased before the end of the grant, and data will be safely kept for 10 years from purchase. If a longer term of storage is needed, PIs may contact Grants and Contracts for approval. Large datasets that are not actively used should also be moved to Bioarchive to save for the higher storage cost on Biocluster or other storage services. | ||
| − | = | + | Any type of data, '''except for the HIPAA data''', is allowed on Bioarchive. |
| − | + | ==How Bioarchive Works== | |
| + | Bioarchive is a disk-to-tape system that utilizes a Spectralogic Black Perl disk system and a Spectralogic T950 tape library with LTO8 tape drives. This setup allows users to transfer data to the archive quickly because data is written to spinning disks, and then the software will move the data to the tape. Unless specified otherwise, all data on Bioarchive is written onto two tapes. One of them stays in the library in case the data is needed, while the other tape is taken to a secure offsite storage facility. | ||
| + | ==How Bioarchive Billing Works== | ||
| + | Each private storage unit is called a Bucket (like a folder), and each bucket needs to be associated with a CFOP number. A user is granted 50GB of storage space before any charge. This is to ensure that the service meets their needs before commitment. Once a bucket passes this initial 50GB threshold, we bill $200 for each TB of data in the next monthly billing period. Ten years after a Bucket is billed, the PI will be offered the choice of removing the data or paying for an additional 10 years. | ||
| − | = | + | To check your Bioarchive billing, go to https://www-app.igb.illinois.edu/bioarchive. if you need to update the CFOP associated with your account, email help@igb.illinois.edu. |
| − | + | =How do I start using Bioarchive= | |
| + | Please fill out the Bioarchive Bucket Request Form: https://www.igb.illinois.edu/webform/archive_bucket_request_form | ||
| + | You will receive an email from CNRG once your account is created. | ||
| + | ==Reset Your Password== | ||
| + | The initial email from CNRG includes a temporary password. If you need us to reset your password, please email [mailto:help@igb.illinois.edu help@igb.illinois.edu] and we can send you a temporary password. | ||
| + | Change your password '''IMMEDIATELY'''. To do so, go to https://bioarchive-login.igb.illinois.edu and log in using your NetID and the temporary password.<div class="mw-parser-output"> | ||
| + | * Click on your NetID at the top-right corner and then choose "User Profile". | ||
| + | * Click on "Action" at the top-left corner and then choose "Edit". | ||
| + | * Enter your temporary password in the Current Password field, and then create a new password and confirm it. Be sure to click "Save" when you are done. | ||
| + | </div> | ||
| − | =Get S3 Keys= | + | ==Get S3 Keys== |
| − | + | The credentials to access your data are the S3 keys that are assigned to your account. To obtain the keys: | |
| − | * Log into | + | * Log into https://bioarchive-login.igb.illinois.edu with the username and password created in the previous step, or continue from there if you just changed your password. |
| − | * | + | * Click on "Action" at the top-left corner and then choose "Show S3 Credentials". |
| − | + | * A pop-up box will appear with your S3 Access ID and S3 Secret Key. Copy them to a safe place as you will need them to access Bioarchive. | |
| − | * A pop-up box will appear with your S3 Access ID and S3 Secret Key | ||
| − | = | + | =How to transfer data to Bioarchive= |
| − | + | We support data transfer from your local computer system and from Biocluster. To prepare for data transfer, you are recommended to put all data for one project in a directory and compress it into a file. Add a readme file to explain the contents inside. Put both the compressed and readme files in a folder. | |
| − | + | ==Data transfer from a computer== | |
| + | The easiest way is to use Eon Browser. Download and install the Eon Browser GUI on your computer. | ||
| + | |||
| + | Windows - [https://help.igb.illinois.edu/images/6/67/BlackPearlEonBrowserSetup-5.0.12.msi BlackPearlEonBrowserSetup-5.0.12.msi] | ||
| + | |||
| + | MacOS - [https://help.igb.illinois.edu/images/6/64/BlackPearlEonBrowser-5.0.12.dmg BlackPearlEonBrowser-5.0.12.dmg] | ||
| − | |||
| − | |||
| − | |||
'''Note for OSX systems''' | '''Note for OSX systems''' | ||
| + | If the system says that the file is corrupted, open a terminal session after you copy the program to your applications. Type the following to the terminal. | ||
| + | <code class="mw-code" style="display:block">cd /Applications | ||
| + | sudo xattr -cr BlackPearlEonBrowser.app | ||
| + | </code> | ||
| + | * If that does not work, please navigate to System Settings>Privacy & Security. Once there, scroll down and you should see a button to allow Eon Browser to open. Click allow and then try to open the program again. This time, you will see the same error message, but it will still give you the option to open the software anyway. | ||
| − | |||
| − | + | ===Using Eon Browser=== | |
| − | + | The Spectralogic User Guide for the Eon Browser is at https://developer.spectralogic.com/wp-content/uploads/2018/11/90990126_C_BlackPearlEonBrowser-UserGuide.pdf.[[File:Screenshot 2024-01-22 114632.png|500px|right|middle|thumb]]<div class="mw-parser-output"> | |
| − | |||
| − | < | ||
| − | + | Use this information when opening a new session on the archive: | |
| + | * Name: Bioarchive | ||
| + | * Data Path Address: bioarchive.igb.illinois.edu | ||
| + | * Port: 443 | ||
| + | * Check the box for SSL | ||
| + | * Access ID: '''Your S3 Access ID''' | ||
| + | * Secret Key: '''Your S3 Secret Key''' | ||
| + | * Check the box for Default Session if you want it to automatically connect to Bioarhive when you open the program. | ||
| + | * Click Save to save your session settings so that future connections will use the same info. | ||
| + | * Click Open to open the archive session. | ||
| − | |||
| − | + | You should see your local computer directories on the left, and your available buckets on the right. To transfer data, just drag from your local computer directories and drop it in the appropriate bucket. You will see the progress in the bottom of the program window. | |
| − | + | When transferring a file, it will first go into the cache and then get written to tape. You can see this happening in the storage locations column. A purple disk icon shows the data is in cache, and the green tape icon shows that the data is on tape. | |
| − | |||
| − | + | While transferring a file, the session will not show a complete transfer until all data is written to the tape. This can take some time. Please check the sleep settings on your computer in advance of the transfer, which can get interrupted if your computer goes to sleep. Using an ethernet instead of a wireless connection would also facilitate data transfer. | |
| − | + | Another way to transfer data from your local system to Bioarchive is to use a command line tool. To do so, download and install the command line tool on your local system: https://developer.spectralogic.com/clients/. The Spectralogic user guide for the command line tool is at https://developer.spectralogic.com/java-command-line-interface-cli-reference-and-examples/. You may need to reconcile your local computer environment with the tool for this to work. Thus, if you aren’t sure about the difference between the two environments, use Eon Browser. The CNRG would not provide further support. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | = | + | ==Data transfer from Biocluster== |
| − | + | The tool to access Bioarchive is already installed on Biocluster. You will need to load the module. | |
| − | + | <code class="mw-code" style="display:block">module load ds3_java_cli | |
| + | </code> | ||
| − | + | This will add the utilities to your environment. Then, load the program we created to set up the environment variables. Enter your secret key and access key when prompted. '''All the bold text in commands below denotes items that need to be replaced with information pertinent to you.''' | |
| − | + | <code class="mw-code" style="display:block">[]$ archive_environment.py | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | < | ||
| − | |||
| − | - | ||
| − | []$ archive_environment.py | ||
Directory exists, but file does not | Directory exists, but file does not | ||
| − | Enter S3 Access ID:S3_Access_ID | + | Enter S3 Access ID: <b>S3_Access_ID</b> |
| − | Enter S3 Secret Key:S3_Secret_Key | + | Enter S3 Secret Key: <b>S3_Secret_Key</b> |
Archive variables already loaded in .bashrc | Archive variables already loaded in .bashrc | ||
Environment updated for next login. To use new values in this session, please type 'source .archive/credentials' | Environment updated for next login. To use new values in this session, please type 'source .archive/credentials' | ||
| − | </ | + | </code> |
| − | + | ||
| − | + | Be sure to type: | |
| − | + | <code class="mw-code" style="display:block">source .archive/credentials | |
| − | + | </code>After your first connection to Bioarchive, you will not be asked for the S3 Access ID and S3 Secret Key again if you connect from the same Biocluster account. | |
| + | |||
| + | |||
| + | To list available buckets: | ||
| + | <code class="mw-code" style="display:block">ds3_java_cli -c get_service | ||
| + | </code>It will only return a list of buckets that the user has permission to access. | ||
| + | |||
| + | |||
| + | To view files in a bucket on Bioarchive: | ||
| + | <code class="mw-code" style="display:block">ds3_java_cli -b <b>Bucket_Name</b> -c get_bucket | ||
| + | </code>This will list all files, their sizes, their owners, and the total size of data in the bucket. | ||
| + | |||
| + | |||
| + | To transfer all the contents in a directory to Bioarchive: | ||
| + | <code class="mw-code" style="display:block">ds3_java_cli -b <b>Bucket_Name</b> -c put_bulk -d <b>Directory_Name</b> | ||
| + | </code>This will transfer everything in this directory, but not the directory itself. | ||
| + | |||
| − | == | + | To transfer a file to Bioarchive: |
| − | + | <code class="mw-code" style="display:block">ds3_java_cli -b <b>Bucket_Name</b> -c put_object -o <b>File_Name</b> | |
| − | + | </code> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | To transfer a file to a certain directory on Bioarchive: |
| − | ds3_java_cli -b | + | <code class="mw-code" style="display:block">ds3_java_cli -b <b>Bucket_Name</b> -c put_object -o <b>File_Name</b> -p <b>Directory/</b> |
| − | + | </code> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | To transfer a file from Bioarchive to Biocluster: | |
| − | ds3_java_cli -b | + | <code class="mw-code" style="display:block">ds3_java_cli -b <b>Bucket_Name</b> -c get_object -o <b>File_Name</b> |
| − | + | </code> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | To transfer all files and directories in a bucket from Bioarchive to Biocluster: | |
| − | + | <code class="mw-code" style="display:block">ds3_java_cli -b <b>Bucket_Name</b> -c get_bulk | |
| − | + | </code> | |
| − | == | + | You can add a -p tag to restrict the transfer scope to a directory. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | <h1>References</h1> | |
| − | |||
| − | |||
| − | |||
| − | + | https://developer.spectralogic.com/java-command-line-interface-cli-reference-and-examples/</div> | |
| − | |||
Revision as of 06:59, 13 September 2024
Contents
Bioarchive[edit]
Bioarchive is set up for long-term data storage at a low upfront cost, which allows grant PIs to preserve data well beyond the funding period. Storage space can be purchased before the end of the grant, and data will be safely kept for 10 years from purchase. If a longer term of storage is needed, PIs may contact Grants and Contracts for approval. Large datasets that are not actively used should also be moved to Bioarchive to save for the higher storage cost on Biocluster or other storage services.
Any type of data, except for the HIPAA data, is allowed on Bioarchive.
How Bioarchive Works[edit]
Bioarchive is a disk-to-tape system that utilizes a Spectralogic Black Perl disk system and a Spectralogic T950 tape library with LTO8 tape drives. This setup allows users to transfer data to the archive quickly because data is written to spinning disks, and then the software will move the data to the tape. Unless specified otherwise, all data on Bioarchive is written onto two tapes. One of them stays in the library in case the data is needed, while the other tape is taken to a secure offsite storage facility.
How Bioarchive Billing Works[edit]
Each private storage unit is called a Bucket (like a folder), and each bucket needs to be associated with a CFOP number. A user is granted 50GB of storage space before any charge. This is to ensure that the service meets their needs before commitment. Once a bucket passes this initial 50GB threshold, we bill $200 for each TB of data in the next monthly billing period. Ten years after a Bucket is billed, the PI will be offered the choice of removing the data or paying for an additional 10 years.
To check your Bioarchive billing, go to https://www-app.igb.illinois.edu/bioarchive. if you need to update the CFOP associated with your account, email help@igb.illinois.edu.
How do I start using Bioarchive[edit]
Please fill out the Bioarchive Bucket Request Form: https://www.igb.illinois.edu/webform/archive_bucket_request_form You will receive an email from CNRG once your account is created.
Reset Your Password[edit]
The initial email from CNRG includes a temporary password. If you need us to reset your password, please email help@igb.illinois.edu and we can send you a temporary password.
Change your password IMMEDIATELY. To do so, go to https://bioarchive-login.igb.illinois.edu and log in using your NetID and the temporary password.
- Click on your NetID at the top-right corner and then choose "User Profile".
- Click on "Action" at the top-left corner and then choose "Edit".
- Enter your temporary password in the Current Password field, and then create a new password and confirm it. Be sure to click "Save" when you are done.
Get S3 Keys[edit]
The credentials to access your data are the S3 keys that are assigned to your account. To obtain the keys:
- Log into https://bioarchive-login.igb.illinois.edu with the username and password created in the previous step, or continue from there if you just changed your password.
- Click on "Action" at the top-left corner and then choose "Show S3 Credentials".
- A pop-up box will appear with your S3 Access ID and S3 Secret Key. Copy them to a safe place as you will need them to access Bioarchive.
How to transfer data to Bioarchive[edit]
We support data transfer from your local computer system and from Biocluster. To prepare for data transfer, you are recommended to put all data for one project in a directory and compress it into a file. Add a readme file to explain the contents inside. Put both the compressed and readme files in a folder.
Data transfer from a computer[edit]
The easiest way is to use Eon Browser. Download and install the Eon Browser GUI on your computer.
Windows - BlackPearlEonBrowserSetup-5.0.12.msi
MacOS - BlackPearlEonBrowser-5.0.12.dmg
Note for OSX systems
If the system says that the file is corrupted, open a terminal session after you copy the program to your applications. Type the following to the terminal.
cd /Applications
sudo xattr -cr BlackPearlEonBrowser.app
- If that does not work, please navigate to System Settings>Privacy & Security. Once there, scroll down and you should see a button to allow Eon Browser to open. Click allow and then try to open the program again. This time, you will see the same error message, but it will still give you the option to open the software anyway.
Using Eon Browser[edit]
The Spectralogic User Guide for the Eon Browser is at https://developer.spectralogic.com/wp-content/uploads/2018/11/90990126_C_BlackPearlEonBrowser-UserGuide.pdf.
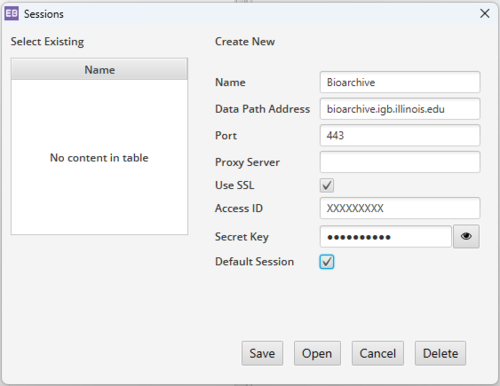
Use this information when opening a new session on the archive:
- Name: Bioarchive
- Data Path Address: bioarchive.igb.illinois.edu
- Port: 443
- Check the box for SSL
- Access ID: Your S3 Access ID
- Secret Key: Your S3 Secret Key
- Check the box for Default Session if you want it to automatically connect to Bioarhive when you open the program.
- Click Save to save your session settings so that future connections will use the same info.
- Click Open to open the archive session.
You should see your local computer directories on the left, and your available buckets on the right. To transfer data, just drag from your local computer directories and drop it in the appropriate bucket. You will see the progress in the bottom of the program window.
When transferring a file, it will first go into the cache and then get written to tape. You can see this happening in the storage locations column. A purple disk icon shows the data is in cache, and the green tape icon shows that the data is on tape.
While transferring a file, the session will not show a complete transfer until all data is written to the tape. This can take some time. Please check the sleep settings on your computer in advance of the transfer, which can get interrupted if your computer goes to sleep. Using an ethernet instead of a wireless connection would also facilitate data transfer.
Another way to transfer data from your local system to Bioarchive is to use a command line tool. To do so, download and install the command line tool on your local system: https://developer.spectralogic.com/clients/. The Spectralogic user guide for the command line tool is at https://developer.spectralogic.com/java-command-line-interface-cli-reference-and-examples/. You may need to reconcile your local computer environment with the tool for this to work. Thus, if you aren’t sure about the difference between the two environments, use Eon Browser. The CNRG would not provide further support.
Data transfer from Biocluster[edit]
The tool to access Bioarchive is already installed on Biocluster. You will need to load the module.
module load ds3_java_cli
This will add the utilities to your environment. Then, load the program we created to set up the environment variables. Enter your secret key and access key when prompted. All the bold text in commands below denotes items that need to be replaced with information pertinent to you.
[]$ archive_environment.py
Directory exists, but file does not
Enter S3 Access ID: S3_Access_ID
Enter S3 Secret Key: S3_Secret_Key
Archive variables already loaded in .bashrc
Environment updated for next login. To use new values in this session, please type 'source .archive/credentials'
Be sure to type:
source .archive/credentials
After your first connection to Bioarchive, you will not be asked for the S3 Access ID and S3 Secret Key again if you connect from the same Biocluster account.
To list available buckets:
ds3_java_cli -c get_service
It will only return a list of buckets that the user has permission to access.
To view files in a bucket on Bioarchive:
ds3_java_cli -b Bucket_Name -c get_bucket
This will list all files, their sizes, their owners, and the total size of data in the bucket.
To transfer all the contents in a directory to Bioarchive:
ds3_java_cli -b Bucket_Name -c put_bulk -d Directory_Name
This will transfer everything in this directory, but not the directory itself.
To transfer a file to Bioarchive:
ds3_java_cli -b Bucket_Name -c put_object -o File_Name
To transfer a file to a certain directory on Bioarchive:
ds3_java_cli -b Bucket_Name -c put_object -o File_Name -p Directory/
To transfer a file from Bioarchive to Biocluster:
ds3_java_cli -b Bucket_Name -c get_object -o File_Name
To transfer all files and directories in a bucket from Bioarchive to Biocluster:
ds3_java_cli -b Bucket_Name -c get_bulk
You can add a -p tag to restrict the transfer scope to a directory.